Hybrid DMRG-like training of MPS
Contents
Hybrid DMRG-like training of MPS#
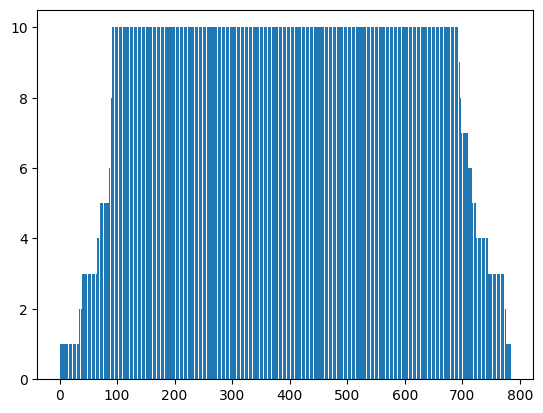
Here we show a way of training MPS models in a DMRG fashion, but where all MPS cores are optimized at the same time, thus making the training process much faster. In this approach, MPS cores are merged in pairs, contracting each node with a neighbour, and the whole model is trained like that. After a few iterations, the cores are unmerged and merged again with the other neighbour. This process can be repeated as many times as desired.
This has the advantage that bond dimensions can be learned during the training process, and also the optimization is much faster than traditional DMRG, since all cores are updated at once.
[ ]:
%mkdir data
%mkdir models
[1]:
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import DataLoader
import torchvision.transforms as transforms
import torchvision.datasets as datasets
import matplotlib.pyplot as plt
import tensorkrowch as tk
[2]:
device = torch.device('cpu')
if torch.cuda.is_available():
device = torch.device('cuda:0')
elif torch.backends.mps.is_available():
device = torch.device('mps:0')
else:
device = torch.device('cpu')
device
[2]:
device(type='cuda', index=0)
Dataset#
[3]:
# MNIST Dataset
dataset_name = 'mnist'
batch_size = 64
image_size = 28
input_size = image_size ** 2
num_classes = 10
transform = transforms.Compose([transforms.ToTensor(),
transforms.Resize(image_size, antialias=True),
])
# Load data
train_dataset = datasets.MNIST(root='data/',
train=True,
transform=transform,
download=True)
test_dataset = datasets.MNIST(root='data/',
train=False,
transform=transform,
download=True)
train_loader = DataLoader(dataset=train_dataset,
batch_size=batch_size,
shuffle=True)
test_loader = DataLoader(dataset=test_dataset,
batch_size=batch_size,
shuffle=True)
[4]:
random_sample = torch.randint(low=0, high=len(train_dataset), size=(1,)).item()
plt.imshow(train_dataset[random_sample][0].squeeze(0), cmap='Greys')
plt.show()
print(train_dataset[random_sample][1])
9
Define model#
[5]:
class MPS_HDMRG(tk.models.MPSLayer):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.out_node.get_axis('input').name = 'output'
self.block_length = None
self.even = None
def merge(self, even, block_length):
n_blocks = self.n_features // block_length
if even:
# Leave reamining nodes at the end
mats_env = self.mats_env[:(n_blocks * block_length)]
else:
# Leave remaining nodes at the beggining
mats_env = self.mats_env[(-n_blocks * block_length):]
blocks = []
for i in range(n_blocks):
block_nodes = mats_env[(i * block_length):((i + 1) * block_length)]
block = block_nodes[0]
for node in block_nodes[1:]:
block = tk.contract_between_(block, node)
block = block.parameterize(True)
block.name = f'block_({i})'
blocks.append(block)
if even:
self._mats_env = blocks + self.mats_env[(n_blocks * block_length):]
else:
self._mats_env = self.mats_env[:(-n_blocks * block_length)] + blocks
self.block_length = block_length
self.even = even
def unmerge(self, side='left', rank=None, cum_percentage=None):
n_blocks = self.n_features // self.block_length
if self.even:
# Leave reamining nodes at the end
blocks = self.mats_env[:n_blocks]
else:
# Leave remaining nodes at the beggining
blocks = self.mats_env[-n_blocks:]
mats_env = []
for i in range(n_blocks):
block = blocks[i]
block_nodes = []
for j in range(self.block_length - 1):
node1_axes = block.axes[:2]
node2_axes = block.axes[2:]
node, block = tk.split_(block,
node1_axes,
node2_axes,
side=side,
rank=rank,
cum_percentage=cum_percentage)
block.get_axis('split').name = 'left'
node.get_axis('split').name = 'right'
node.name = f'mats_env_({i * self.block_length + j})'
node = node.parameterize(True)
block_nodes.append(node)
block.name = f'mats_env_({i * self.block_length + j + 1})'
block = block.parameterize(True)
block_nodes.append(block)
mats_env += block_nodes
if self.even:
self._mats_env = mats_env + self.mats_env[n_blocks:]
else:
self._mats_env = self.mats_env[:-n_blocks ] + mats_env
self.block_length = None
self.even = None
def contract(self):
result_mats = []
for node in self.mats_env:
while any(['input' in name for name in node.axes_names]):
for axis in node.axes:
if 'input' in axis.name:
data_node = node.neighbours(axis)
node = node @ data_node
break
result_mats.append(node)
result_mats = [self.left_node] + result_mats + [self.right_node]
result = result_mats[0]
for node in result_mats[1:]:
result @= node
return result
[6]:
# Model hyperparameters
embedding_dim = 3
output_dim = num_classes
bond_dim = 10
init_method = 'randn_eye'
block_length = 2
cum_percentage = 0.98
[7]:
# Initialize network
model_name = 'mps_dmrg_hybrid'
mps = MPS_HDMRG(n_features=input_size + 1,
in_dim=embedding_dim,
out_dim=num_classes,
bond_dim=bond_dim,
boundary='obc',
init_method=init_method,
std=1e-6,
device=device)
# Important to set data nodes before merging nodes
mps.set_data_nodes()
[8]:
def embedding(x):
x = tk.embeddings.poly(x, degree=embedding_dim - 1)
return x
Train#
[9]:
# Hyperparameters
learning_rate = 1e-4
weight_decay = 1e-8
num_epochs = 10
move_block_epochs = 100
# Loss and optimizer
criterion = nn.CrossEntropyLoss()
[10]:
# Check accuracy on training & test to see how good our model is
def check_accuracy(loader, model):
num_correct = 0
num_samples = 0
model.eval()
with torch.no_grad():
for x, y in loader:
x = x.to(device)
y = y.to(device)
x = x.reshape(x.shape[0], -1)
scores = model(embedding(x))
_, predictions = scores.max(1)
num_correct += (predictions == y).sum()
num_samples += predictions.size(0)
accuracy = float(num_correct) / float(num_samples) * 100
model.train()
return accuracy
[11]:
# Train network
even = True
mps.merge(even, block_length)
mps.trace(torch.zeros(1, input_size, embedding_dim, device=device))
optimizer = optim.Adam(mps.parameters(),
lr=learning_rate,
weight_decay=weight_decay)
for epoch in range(num_epochs):
for batch_idx, (data, targets) in enumerate(train_loader):
# Get data to cuda if possible
data = data.to(device)
targets = targets.to(device)
# Get to correct shape
data = data.reshape(data.shape[0], -1)
# Forward
scores = mps(embedding(data))
loss = criterion(scores, targets)
# Backward
optimizer.zero_grad()
loss.backward()
# Gradient descent
optimizer.step()
if (batch_idx + 1) % move_block_epochs == 0:
if even:
mps.unmerge(side='left',
rank=bond_dim,
cum_percentage=cum_percentage)
else:
mps.unmerge(side='right',
rank=bond_dim,
cum_percentage=cum_percentage)
even = not even
mps.merge(even, block_length)
mps.trace(torch.zeros(1, input_size, embedding_dim, device=device))
optimizer = optim.Adam(mps.parameters(),
lr=learning_rate,
weight_decay=weight_decay)
train_acc = check_accuracy(train_loader, mps)
test_acc = check_accuracy(test_loader, mps)
print(f'* Epoch {epoch + 1:<3} ({even=}) => Train. Acc.: {train_acc:.2f},'
f' Test Acc.: {test_acc:.2f}')
# Reset before saving the model
mps.reset()
torch.save(mps.state_dict(), f'models/{model_name}_{dataset_name}.pt')
* Epoch 1 (even=False) => Train. Acc.: 95.27, Test Acc.: 95.25
* Epoch 2 (even=True) => Train. Acc.: 96.49, Test Acc.: 96.24
* Epoch 3 (even=False) => Train. Acc.: 97.59, Test Acc.: 96.84
* Epoch 4 (even=True) => Train. Acc.: 97.81, Test Acc.: 97.15
* Epoch 5 (even=False) => Train. Acc.: 98.38, Test Acc.: 97.52
* Epoch 6 (even=True) => Train. Acc.: 98.29, Test Acc.: 97.65
* Epoch 7 (even=False) => Train. Acc.: 98.38, Test Acc.: 97.70
* Epoch 8 (even=True) => Train. Acc.: 98.45, Test Acc.: 97.62
* Epoch 9 (even=False) => Train. Acc.: 98.42, Test Acc.: 97.69
* Epoch 10 (even=True) => Train. Acc.: 98.37, Test Acc.: 97.24
[14]:
mps.unmerge(rank=bond_dim, cum_percentage=cum_percentage)
[15]:
mps.update_bond_dim()
[16]:
plt.bar(torch.arange(mps.n_features - 1) + 1, torch.tensor(mps.bond_dim))
plt.show()